Within chemical biology, covalent chemical probes play an important role – they can be used to label proteins according to a certain trait, which include post-translational modifications (PTMs), an enzymatic activity or the affinity towards a drug. Synthetic chemistry allows the synthesis of broad-spectrum probes or probes with very specific reactivity. A variety of read-outs are possible through the use of different chemical handles (biotin, fluorophores, bioorthogonal functional groups). Identification of targets is possible by proteomics methods.
Our laboratory develops and applies small molecule tools for peptidases (often referred to as proteases), with a focus on serine and cysteine proteases, and intramembrane proteases.
Proteases and ABPs
Proteases are enzymes responsible for the cleavage of peptide bonds. They are not only involved in digestive processes and protein turnover, but also play a crucial role in regulatory processes, such as apoptosis, blood coagulation and antigen presentation. Therefore, it is not surprising that misregulation of proteases can lead to a wide variety of diseases.
Proteases are generally translated as inactive zymogens and need proteolytic processing to become active. Once active, a variety of regulatory mechanisms exist to keep protease activity in check – for example, PTMs, the presence of endogenous inhibitors, and the degradation of proteases (Figure 1A). However, only the active protease species is able to convert a substate to a product, which leads to a downstream effect.
To specifically label the active form of proteases, we develop and use activity-based probes (ABPs – Figure 1B). These are often derived from electrophilic inhibitors that react in a mechanism-based manner with the active site machinery. This reaction leads to covalent attachment of a detection tag, which can be used for visualisation, analysis or enrichment of the active protease species. For their synthesis we make use of different solution and solid phase chemistries. The latter is preferred, since they can be easily carried out and facilitate optimisation for different protease targets.
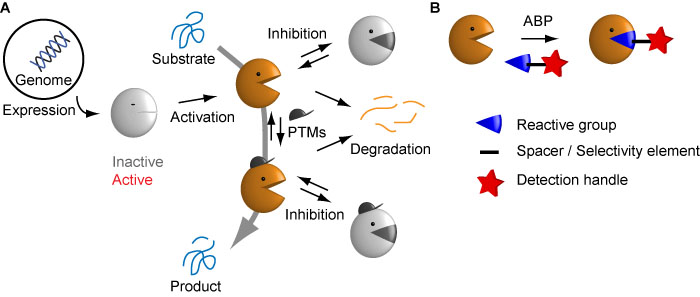
Figure 1. A) The complex biochemistry of proteases. Proteases are synthesized as inactive forms, and active forms are tightly regulated by post-translational mechanisms. Naturally, only the active forms are responsible for biological effects. B) Activity-based probes specifically detect active proteases by means of a reactive group that engages in a mechanism-based reaction with the active site. Figure adapted from Verdoes & Verhelst, Biochem Biophys Acta 2015
Intramembrane proteases
Intramembrane proteases (IMPs) cleave their substrates, which are also membrane proteins in a transmembrane helix (TM) or a juxtamembrane region. Through this cleavage, a soluble part of the substrate is released from the membrane into the exterior or interior of the cell or organelle, where it can function in downstream pathways, in a role distinct from the full-length progenitor form. Rhomboid proteases occur in virtually all sequenced organisms and are involved in a wide variety of biologically important and medically relevant processes. They utilize a catalytic dyad of serine and histidine, located aproximately 10 Å below the surface of the membrane (Figure 2A).
We have developed several activity assays for rhomboid, using either MALDI mass spectrometry or ABPs (Figure 2B-D). With these assays, we have discovered novel inhibitors and ABPs. The use of inhibitors and probes has led to novel insights in the substrate binding mechanism and in the possitble use of probes for high throughput screening and fluorescent microscopy. We continue to develop inhibitors and probes in order to elucidate the functional roles and biochemical mechanism of this fascinating family of enzymes.
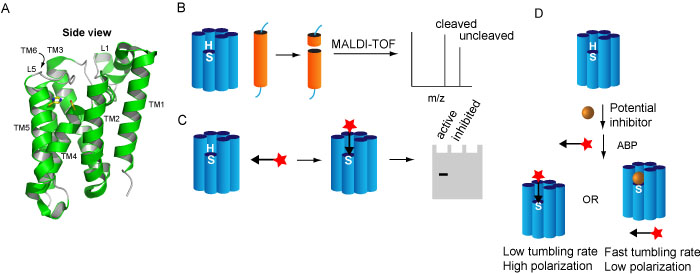
Figure 2. A) Crystal structure of a rhomboid protease. B-D) different activity assays for rhomboids, using MALDI-TOF (B), gel-mediated activity-based protein profiling (C) or fluorescent polarization activity-based protein profiling (D).
Cleavable linkers
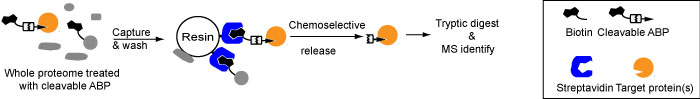
A crucial part in chemical biology is the identification of the protein targets of covalent chemical probes. We have started a program to develop cleavable linkers in order to identify these protein targets with lower false positives and with the aim of mapping the precise binding site (Figure 3).